Welcome to ALL-IDB initiative
We propose a new public and free dataset of microscopic images of blood samples, specifically designed for the evaluation and the comparison of algorithms for segmentation and image classification. The initiative is focused on Acute Lymphoblastic Leukemia (ALL), a serious blood pathology that can being fatal in as little as a few weeks if left untreated, most common in childhood with a peak incidence at 2-5 years of age.
For each image in the dataset, the classification/position of ALL lymphoblasts is provided by expert oncologists. Furthermore, we suggest a specific set of figure of merits to be processed in order to fairly compare different algorithms with the proposed dataset.
We hope that this initiative could give a new test tool to the image processing and pattern matching communities, aiming to stimulate new studies in this important field of research.
The datasets
The images of the dataset has been captured with an optical laboratory microscope coupled with a Canon PowerShot G5 camera. All images are in JPG format with 24 bit color depth, resolution 2592 x 1944.
Dataset ALL_IDB1
The ALL_IDB1 version 1.0 can be used both for testing segmentation capability of algorithms, as well as the classification systems and image preprocessing methods. This dataset is composed of 108 images collected during September, 2005. It contains about 39000 blood elements, where the lymphocytes has been labeled by expert oncologists. The images are taken with different magnifications of the microscope ranging from 300 to 500.
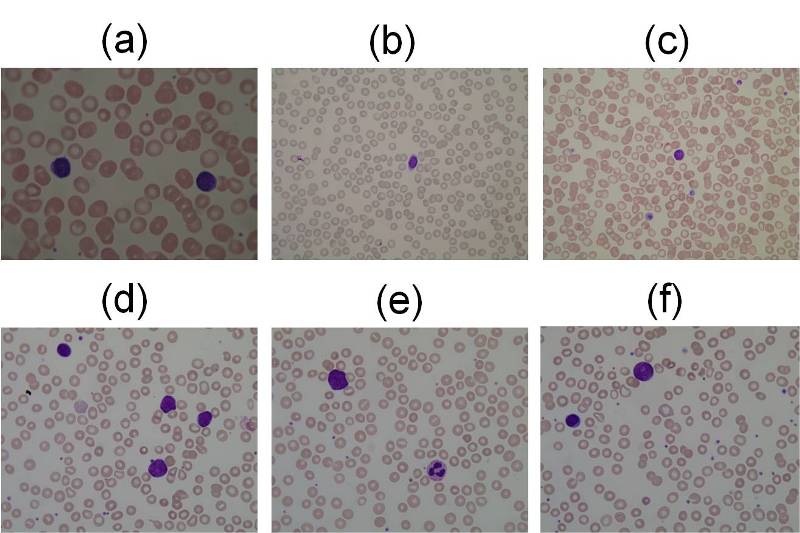
Examples of the images contained in ALL-IDB1: healthy cells from non-ALL patients (a-c),
probable lymphoblasts from ALL patients (d-f).
(a)
''File Im006_1.jpg''
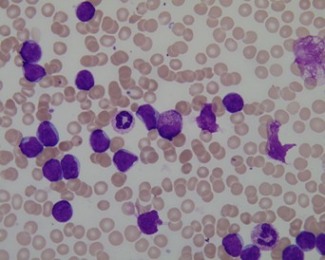
(b)
Content of file "Im006_1.xyc"
446
164
168
442
248
...
62
279
377
415
713
...
Example of the ALL-IDB1 annotation: input image ''Im006_1.jpg'' (a) and the related classification file ''Im006_1.xyc'' reporting the coordinates of the centroids of probable ALL lymphoblasts (b).
The annotation of ALL-IDB1 is as follows. The ALL-IDB1 image files are named with the notation ImXXX_Y.jpg where XXX is a 3-digit integer counter and Y is a boolean digit equal to 0 is no blast cells are present, and equal to 1 if at least one blast cell is present in the image. Please note that all images labeled with Y=0 are from for healthy individuals, and all images labeled with Y=1 are from ALL patients. Each image file ImXXX_Y.jpg is associated with a text file ImXXX_Y.xyc reporting the coordinates of the centroids of the blast cells, if any.
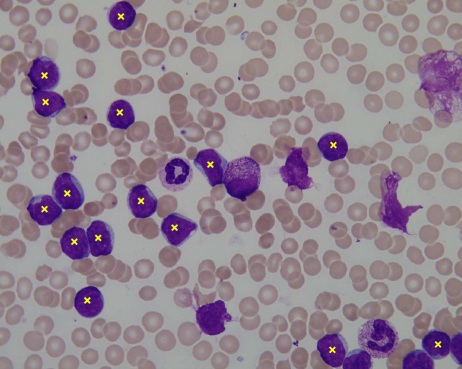
Visualization of the classification data. The centroids stored in the ''Im006_1.xyc'' are plotted on the input image ''Im006_1.jpg'' as crosses to indicate the position of probable ALL lymphoblasts.
ALL-IDB1 allows for different levels of analysis:
| Algorithm type and description | Pseudocode | Verification |
|---|---|---|
| Image classification The algorithm estimates if the input image is coming from a ALL patient. |
CLASS = classifier ( "ImXXX_Y.jpg" ); |
The image classification is correct if and only if the output CLASS is equal to Y Alternative test: the image classification is correct if and only if the output CLASS is equal to boolean Z = M>0, where M is the number of rows in the ImXXX_Y.xyc file. |
| ALL Blast cell counter The algorithm estimates the number of blast cells in the input image (if output > 0, --> ALL patient). |
N = blastCounter ( "ImXXX_Y.jpg"); |
the output N is correct if equal to the M rows in the ImXXX_Y.xyc file |
| ALL Blast cell identifier The algorithm estimates the location of the centroids of the blast cells in the input image (if the output cardinality is > 0 --> ALL patient). |
COORDINATES = blastIdentifier ( "ImXXX_Y.jpg" ); |
Error = accuracy(COORDINATES , "ImXXX_Y.xyc" ); e.g., function accuracy returns the number of correct matches (for example, within a 10 pixel radius) in the COORDINATES list. |
Dataset ALL_IDB2
This image set has been designed for testing the performances of classification systems. The ALL-IDB2 version 1.0 is a collection of cropped area of interest of normal and blast cells that belongs to the ALL-IDB1 dataset. ALL-IDB2 images have similar gray level properties to the images of the ALL-IDB1, except the image dimensions.
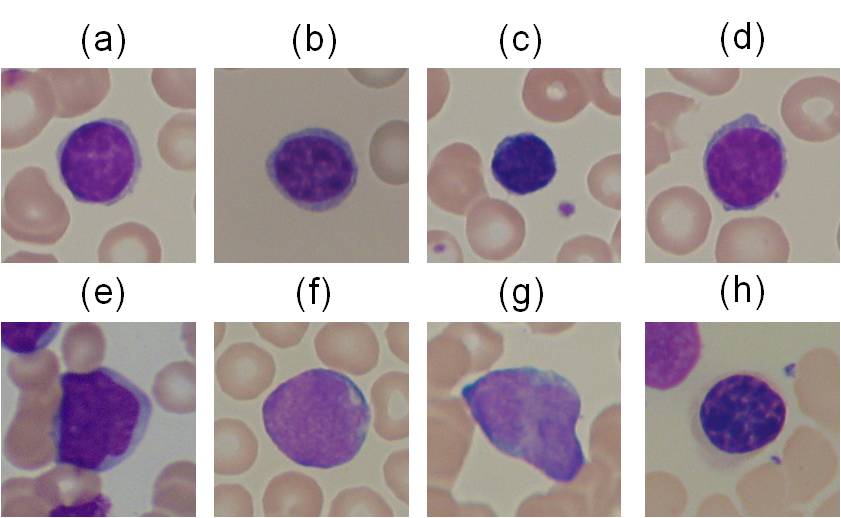
Examples of the images contained in ALL-IDB2: healthy cells from non-ALL patients (a-d),
probable lymphoblasts from ALL patients (e-h).
The annotation of ALL-IDB2 is as follows. The ALL-IDB2 image files are named with the notation ImXXX_Y.jpg where XXX is a progressive 3-digit integer and Y is a boolean digit equal to 0 if the cell placed in the center of the image is not a blast cell, and equal to 1 if the cell placed in the center of the image is a blast cell. Please note that all images labeled with Y=0 are from for healthy individuals, and all images labeled with Y=1 are from ALL patients.
ALL-IDB2 allows for the following level of analysis.
| Algorithm type and description | Pseudocode | Verification |
|---|---|---|
| Image classification The algorithm estimates if the input image is coming from a ALL patient. |
CLASS = classifier ( "ImXXX_Y.jpg" ); |
The image classification is correct if and only if the output CLASS is equal to Y |
Background
Morphological features of ALL blast cells
The classification of the lymphocyte in microscope images is quite complex since even an expert operator can have dubs in classifying some lymphocyte cells. Actually, the morphological distinctive aspects of ALL blast and normal lymphocytes are very smooth.
Of course, nowadays more accurate diagnostic tools area available (e.g., the immunologic classification) but they require a blood sample and, since they are not image-based, their usage in telemedicine applications is quite limited. According to the most common visual morphological analysis for the ALL disease (the FAB method, see related publications), the features that trained lab technicians consider during the image observation are the following:
- L1: ALL blasts are small and homogeneous. The nuclei are round and regular with little clefting and inconspicuous nucleoli. Cytoplasm is scanty and usually without vacuoles.
- L2: ALL blasts are large and heterogeneous. The nuclei are irregular and often clefted. One or more, usually large nucleoli are present. The volume of cytoplasm is variable, but often abundant and may contain vacuoles.
- L3: ALL blasts are moderate-large in size and homogeneous. The nuclei are regular and round-oval in shape. One or more prominent nucleoli are present. The volume of cytoplasm is moderate and contains prominent vacuoles.
Next figure shows the great variability in shape and pattern of the blast cells according to the FAB classification for the ALL disease. The main goal is to detect without differentiation the presence of all three types of blasts in the images.
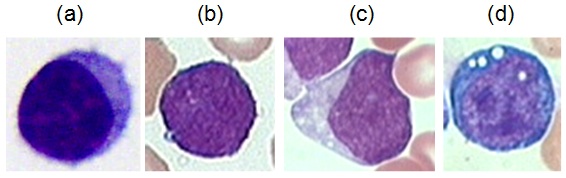
Morphological variability associated to the blast cells according to the FAB classification:
(a) healthy lymphocytes cell from non-ALL patients, (b-d) lymphoblasts from ALL patients where (b), (c) and (d) are L1, L2 and L3 respectively.
Image processing
In the literature, the identification and classification of white blood blast cells had been tackled by a classic sequence of steps as shown in the next figure. This approach can be followed for ALL-IDB1 and ALL-IDB2.
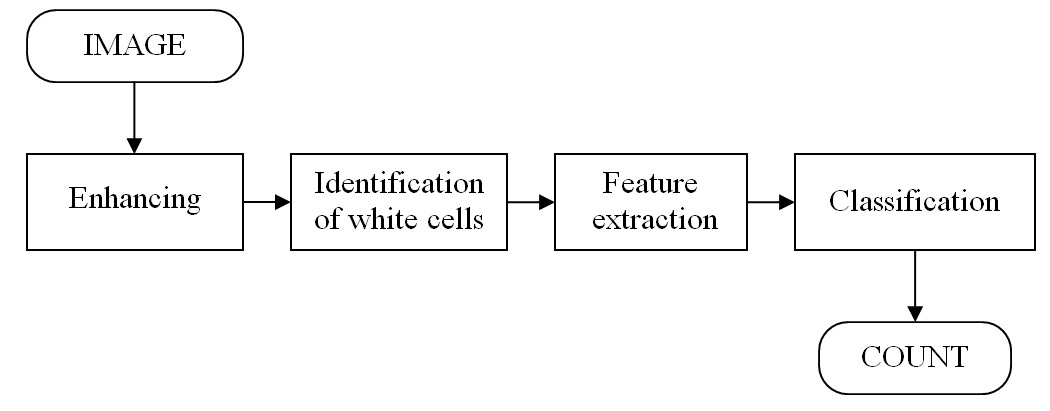
A possible approach to ALL image classification.
More in detail (specifically for the for ALL-IDB1) a hierarchical classification approach can be followed, where the segmentation of white cells is achieved and then each single cell is classified after a feature extraction phase.
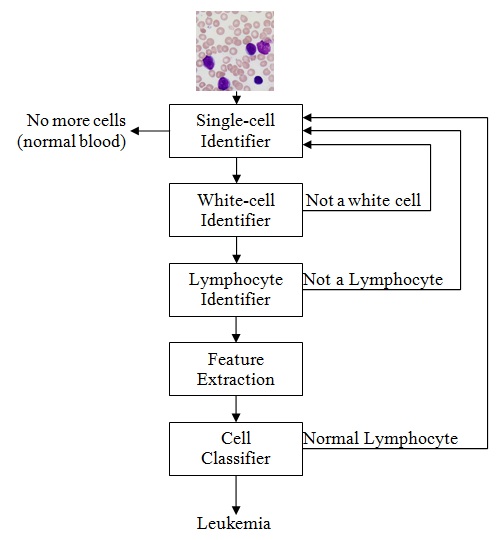
Example of ALL blast cells recognition by segmentation and single cell classification.
If you are interested in image segmentation and automatic morphological analysis, you may find additional details in the following related publications:
- "Robust Segmentation and Measurements Techniques of White Cells in Blood Microscope Images"
- "Automatic morphological analysis for acute leukemia identification in peripheral blood microscope images"
Download and Term of use
Obtaining and using the dataset
All requests for the ALL-IDB datasets must be directed (by email) to Fabio Scotti
( fabio DOT scotti AT unimi DOT it ).
Applicants should manually fill, sign, scan and attach the application form to the given email address.
Upon receipt of an executed copy of the signed application form, access instructions will be given.
Download Application Form (PDF).
Citation
All documents and papers that report on research that uses the ALL-IDB datasets must include an appropriate citation (see the related publications section).
Warning
We strongly discourage the use of the ALL-IDB content for diagnostic or different activities than the purpose of this initiative. ALL-IDB must be considered as an image processing dataset.
Estimating the accuracy of algorithms on ALL-IDB
We propose a specific set of features to be processed in order to fairly compare different algorithms tested on ALL-IDB images. Please consider this section: reporting the results .
Acknowledgements
We wish to thanks prof. Andrea Biondi and Dr. Oscar Maglia from M. Tettamanti Research Center -Monza, Italy- for their profitable cooperation and encouragement, the data provided and the careful classification of sample images. The Tettamanti Research Center is part of the Tettamanti Foundation, a no-profit scientific institution, involved in the area of research on childhood leukemias and hematological diseases, based at the S. Gerardo Hospital in Monza, Italy.
People
The ALL-IDB initiative is sustained and maintained by
- Fabio Scotti (proposer and maintainer)
- Ruggero Donida Labati
- Vincenzo Piuri
Related publications
- R. Donida Labati, V. Piuri, F. Scotti, "ALL-IDB: the acute lymphoblastic leukemia image database for image processing", in Proc. of the 2011 IEEE Int. Conf. on Image Processing (ICIP 2011), Brussels, Belgium, pp. 2045-2048, September 11-14, 2011. ISBN: 978-1-4577-1302-6. [DOI: 10.1109/ICIP.2011.6115881][PDF] [BibTex entry]
- F. Scotti, "Robust Segmentation and Measurements Techniques of White Cells in Blood Microscope Images", in Proc. of the 2006 IEEE Instrumentation and Measurement Technology Conf. (IMTC 2006), Sorrento, Italy, pp. 43-48, April 24-27, 2006. ISSN: 1091-5281. [DOI: 10.1109/IMTC.2006.328170 ][PDF] [BibTex entry]
- F. Scotti, "Automatic morphological analysis for acute leukemia identification in peripheral blood microscope images", in Proc. of the 2005 IEEE Int. Conf. on Computational Intelligence for Measurement Systems and Applications (CIMSA 2005), Giardini Naxos - Taormina, Italy, pp. 96-101, July 20-22, 2005 [DOI: 10.1109/CIMSA.2005.1522835][PDF] [BibTex entry]
- V. Piuri, F. Scotti, "Morphological classification of blood leucocytes by microscope images", in Proc. of the 2004 IEEE Int. Conf. on Computational Intelligence for Measurement Systems and Applications (CIMSA 2004), Boston, MA, USA, pp. 103-108, July 12-14, 2004. ISBN: 0-7803-8341-9. [DOI: 10.1109/CIMSA.2004.1397242][PDF] [BibTex entry]
Related Projects
